biotech
Methods for the study of the tissue microenvironment using spatial statistics
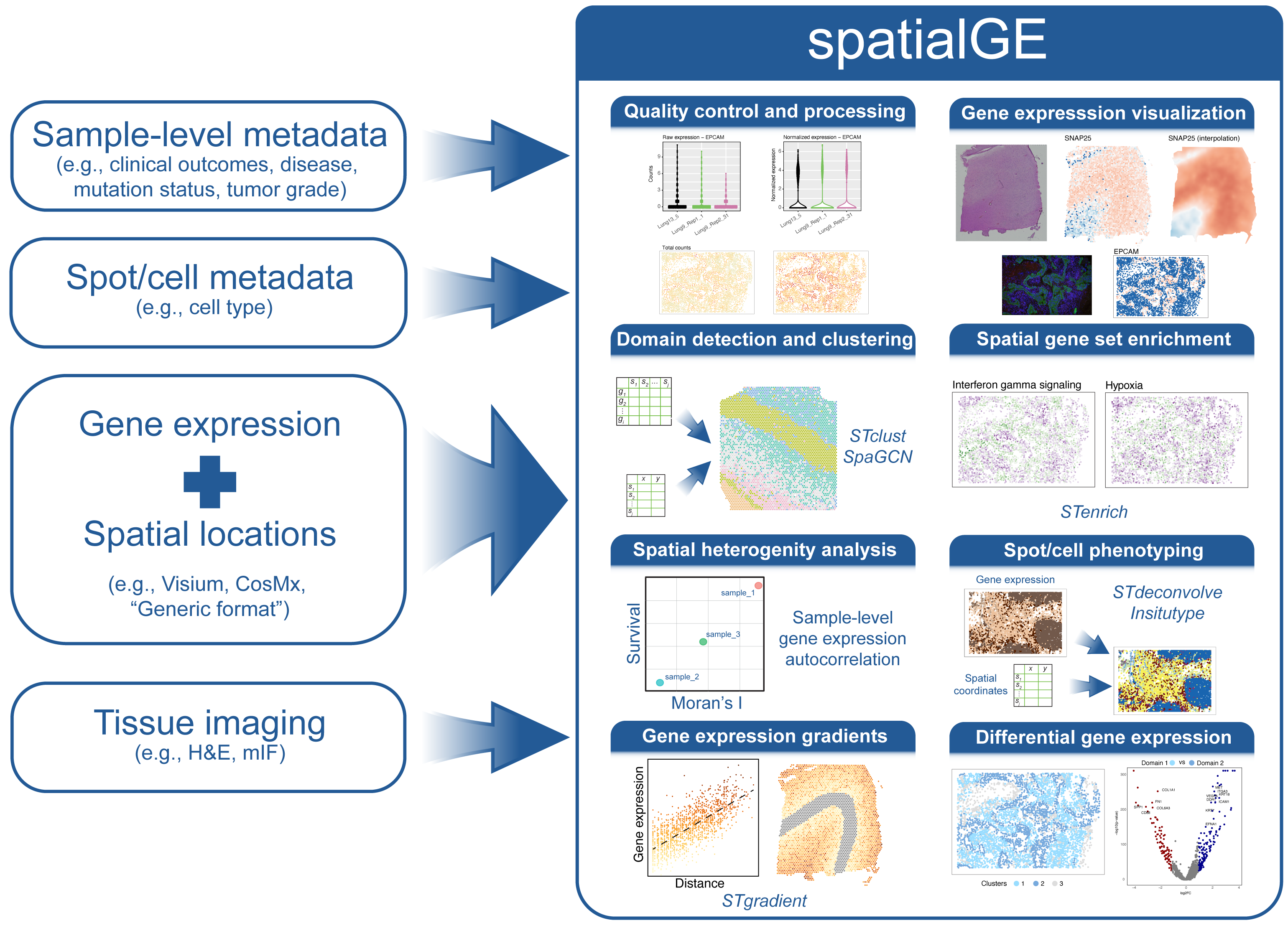
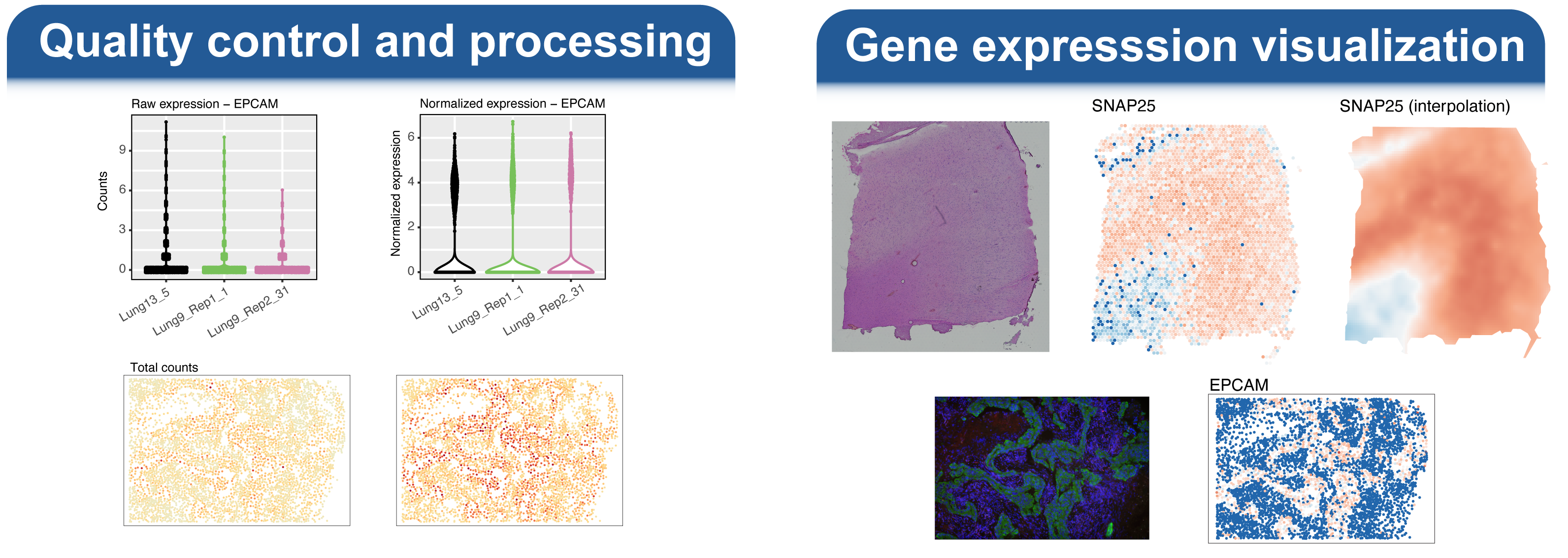
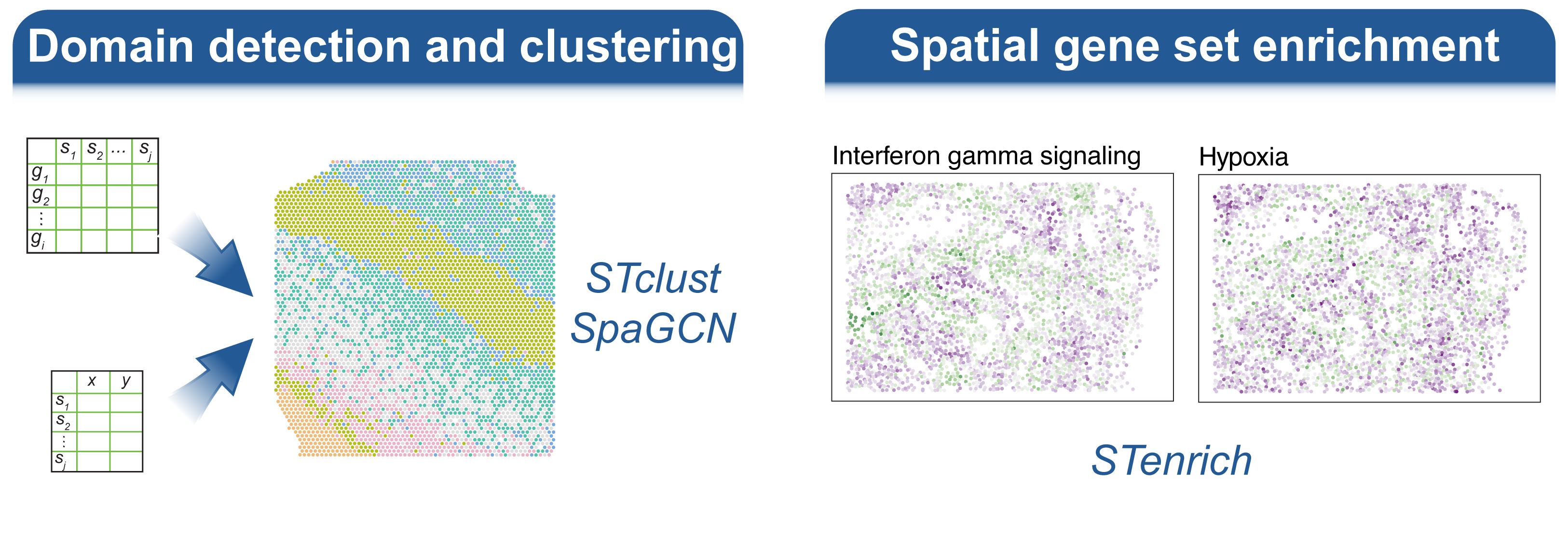
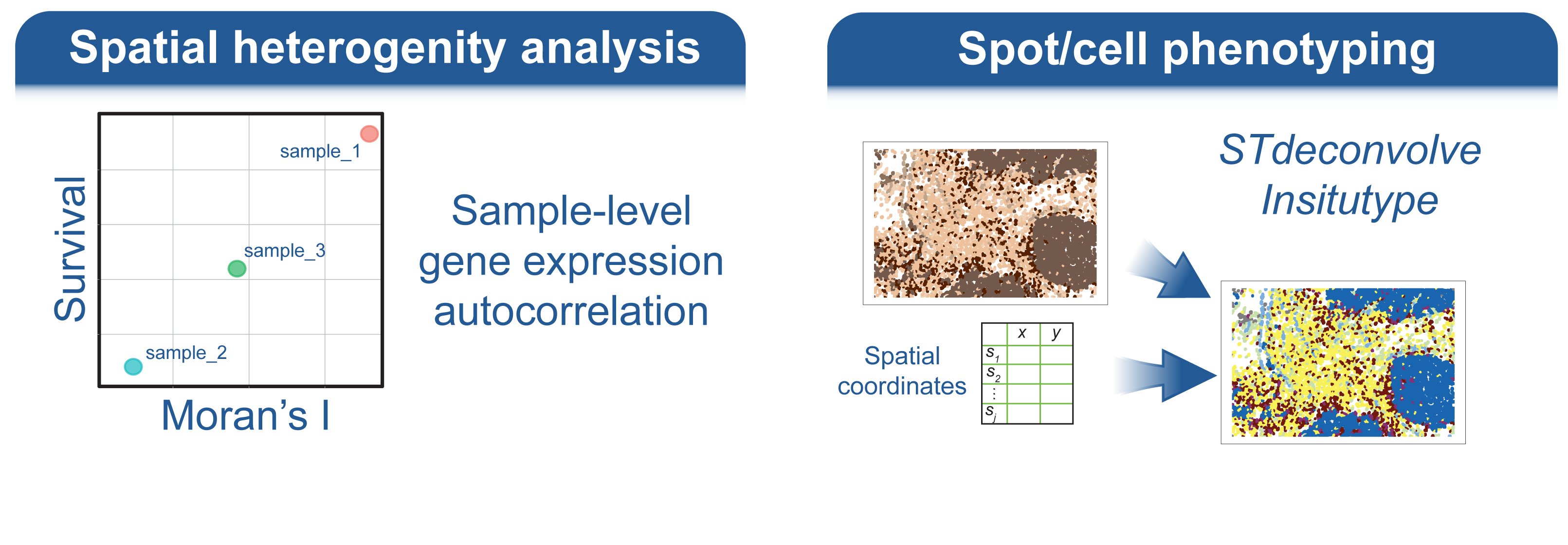
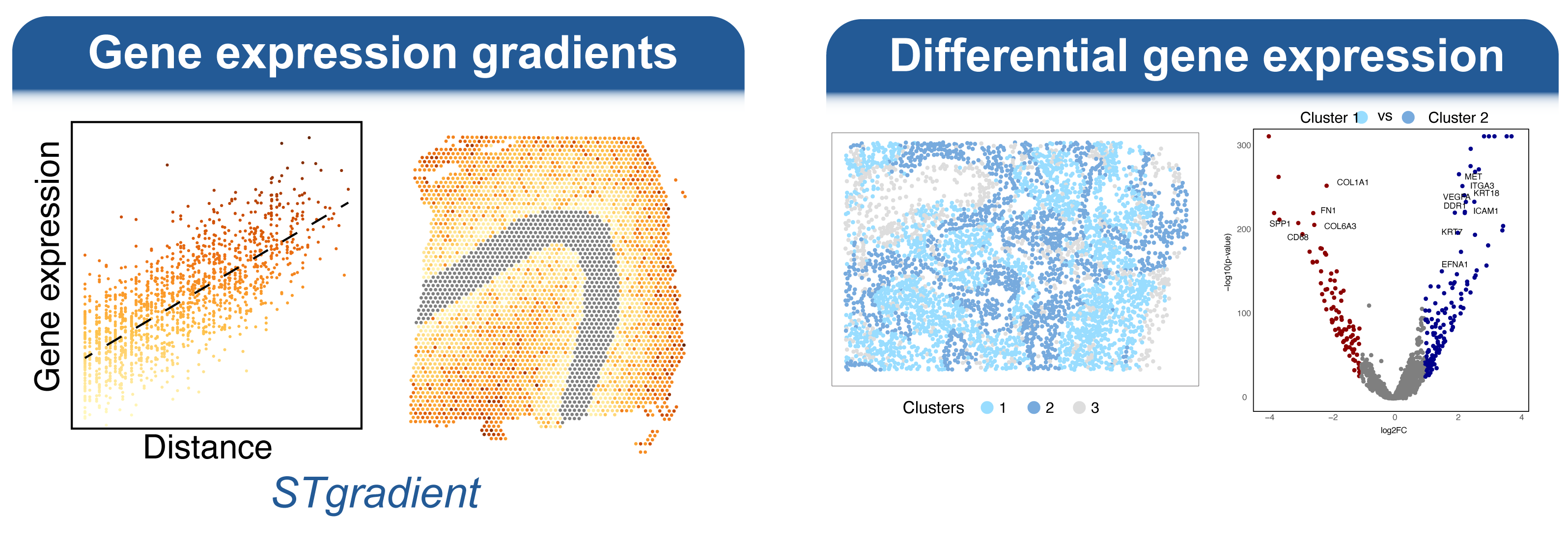
Welcome to the spatialGE web application, a user friendly, point-and-click implementation of the spatialGE R package. This application contains a collection of methods for visualization and spatial statistics analysis of the tissue microenvironment and heterogeneity using spatial transcriptomics (ST) experiments. For a technical description of the methods, please see our publications at the bottom of this page.
For tutorials and guides on how to use spatialGE, please refer to the “How to get started” section, or click here to log in your account.
Publications
If you use spatialGE for analysis you present in a publication or abstract, please cite the following paper:
- Ospina, O, E., Manjarres-Betancur, R., Gonzalez-Calderon, G., Soupir, A. C., Smalley, I., Tsai, K. Y., Markowitz, J., Khaled, M. L., Vallebuona, E., Berglund, A., Eschrich, S., Yu, X. Fridley, B. L. 2025. spatialGE Is a User-Friendly Web Application That Facilitates Spatial Transcriptomics Data Analysis. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-24-2346
Some scientific articles using spatialGE methods:
- Alhaddad, H., Ospina, O. E., Khaled, M. L., Ren, Y., Vallebuona, E., Boozo, M. B., Forsyth, P. A., Pina, Y., Macaulay, R., Law, V., Tsai, K. Y., Cress, W. D., Fridley, B. L., Smalley, I. 2024. Spatial transcriptomics analysis identifies a tumor-promoting function of the meningeal stroma in melanoma leptomeningeal disease. Cell Reports Medicine 5: 101606. https://doi.org/10.1016/j.xcrm.2024.101606
- Soupir, A. C., Hayes, M. T., Peak, T. C., Ospina, O. E., Chakiryan, N. H., Berglund, A., Stewart, P. A., Nguyen, J. V., Moran-Segura, C. M., Francis, N. L., Ramos-Echevarria, P. M., Chahoud, J., Li, R., Tsai, K. Y., Balasi, J. A., Caraballo-Perez, Y., Dhillon, J., Martinez, L. A., Gloria, W. E., Schurman, N., Kim, S., Gregory, M., Mule, J. J., Fridley, B. L., Manley, B. J. 2023. Increased spatial coupling of integrin and collagen IV in the immunoresistant clear cell renal cell carcinoma tumor microenvironment. bioRxiv. https://www.biorxiv.org/content/10.1101/2023.11.16.567457v1
- Ospina, O. E., Soupir, A. C., Fridley, B. L. 2023. A primer on preprocessing, visualization, clustering, and phenotyping of barcode-based spatial transcriptomics data. In: Fridley, B. L., Wang, X. (eds) Statistical Genomics. Methods in Molecular Biology, New York, NY, USA. https://doi.org/10.1007/978-1-0716-2986-4_7