Frequently Asked Questions
I have data from a GeoMx experiment. Can I analyze it with spatialGE?
Yes, you can. spatialGE can be applied to any spatially-resolved transcriptomics experiment as long as gene counts are associated with x and y coordinates within a tissue sample. Some of the methods in spatialGE have been designed to analyze spatial data that is densely sampled, and most GeoMx experiments are characterized by ROI-based sampling that often is sparse and targeting tissue compartments. As a result, some methods would not yield insightful results. For example, while creating a gene expression surface (interpolation) based on 20 ROIs across a large tissue section is possible, it would likely not be informative.
Do I have to pay to use spatialGE?
No, spatialGE is open to the research community. Development of spatialGE has been funded by NIH grants (T32 CA 233399 and U01 CA 274489). Users are only required to create an account, and to acknowledge its use by citing the research articles that describes the methods in spatialGE:
Ospina, O, E., Manjarres-Betancur, R., Gonzalez-Calderon, G., Soupir, A. C., Smalley, I., Tsai, K. Y., Markowitz, J., Khaled, M. L., Vallebuona, E., Berglund, A., Eschrich, S., Yu, X. Fridley, B. L. 2024. spatialGE: A user-friendly web application to democratize spatial transcriptomics analysis. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-24-2346
Ospina, O. E., Wilson, C. M., Soupir, A. C., Berglund, A., Smalley, I., Tsai, K. Y., Fridley, B. L. 2022. spatialGE: Quantification and visualization of the tumor microenvironment heterogeneity using spatial transcriptomics. Bioinformatics 38: 2645–2647. https://doi.org/10.1093/bioinformatics/btac145
What does spatialGE do with my data?
Any sample data you upload to spatialGE can only be accessed using your credentials. Please, do not upload any PHI.
How long is my data stored in spatialGE?
spatialGE stores data at the project level. Multiple projects can be created and kept within the user’s account. There is a 1-year inactive account policy in place. When a project is not used or accessed for a period of one year, the project is deleted (including all data and files), but users will be notified via email 3 months and 1 month before.
Is it normal for some methods in spatialGE to take long time to complete?
Some methods implemented in spatialGE are memory and computing intensive (e.g., spatial differential expression, gene expression surface generation). The methods in spatialGE use R code to conduct analyses (code available here), which could be downloaded and installed to use via command-line requiring basic knowledge of R programming. Should the user choose to use the spatialGE web app, when running an analysis, the user is prompted to select to receive an email notifying when an analysis is complete. Once the analysis is started and an email notification selected, the user can close spatialGE and the process will keep running.
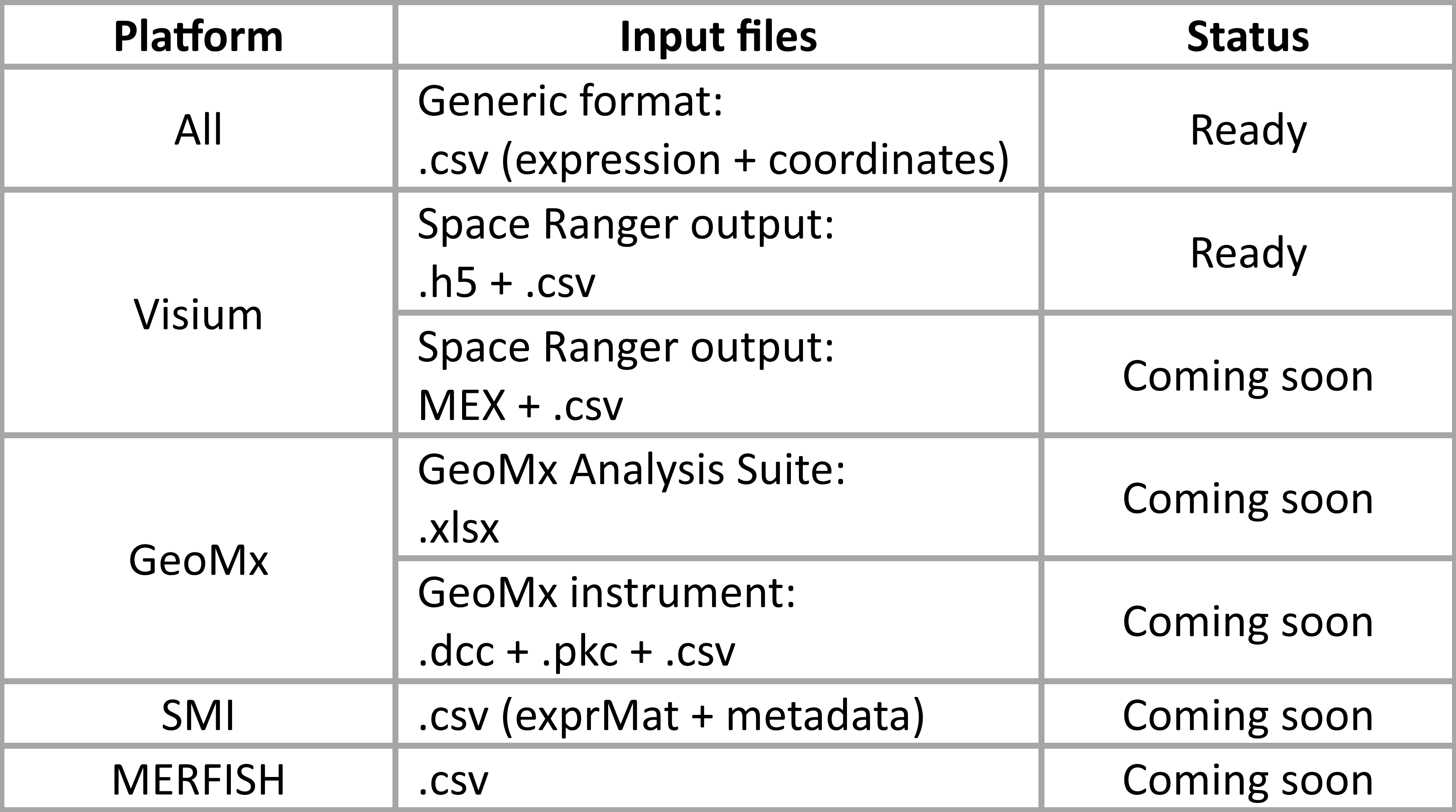
Which spatial transcriptomics formats can I analyze in spatialGE?
The spatialGE web application currently supports Visium data sets. Many more formats will be available soon (see table below). If you would like to analyze other spatial transcriptiomics data formats, please use the command-line R package.
What is the maximum number of samples per project?
While there is not a maximum number of samples per project, the spatialGE web application has been designed as an exploratory tool and projects with more than 10 samples are not recommended. Projects with more than 10 samples might experience long queue and execution times. For analysis of larger projects, the use of the spatialGE R package is recommended, installed in either a personal laptop or a high-performance computing (HPC/cluster) environment.
How many analyses can I run at the same time?
Because spatialGE is a free tool open to the public, you can only run one analysis at a time. A maximum number of users can run analyses concurrently, when that number is reached, any additional analyses will be queued until analyses from other users are completed and enough computing resources for your job are available.